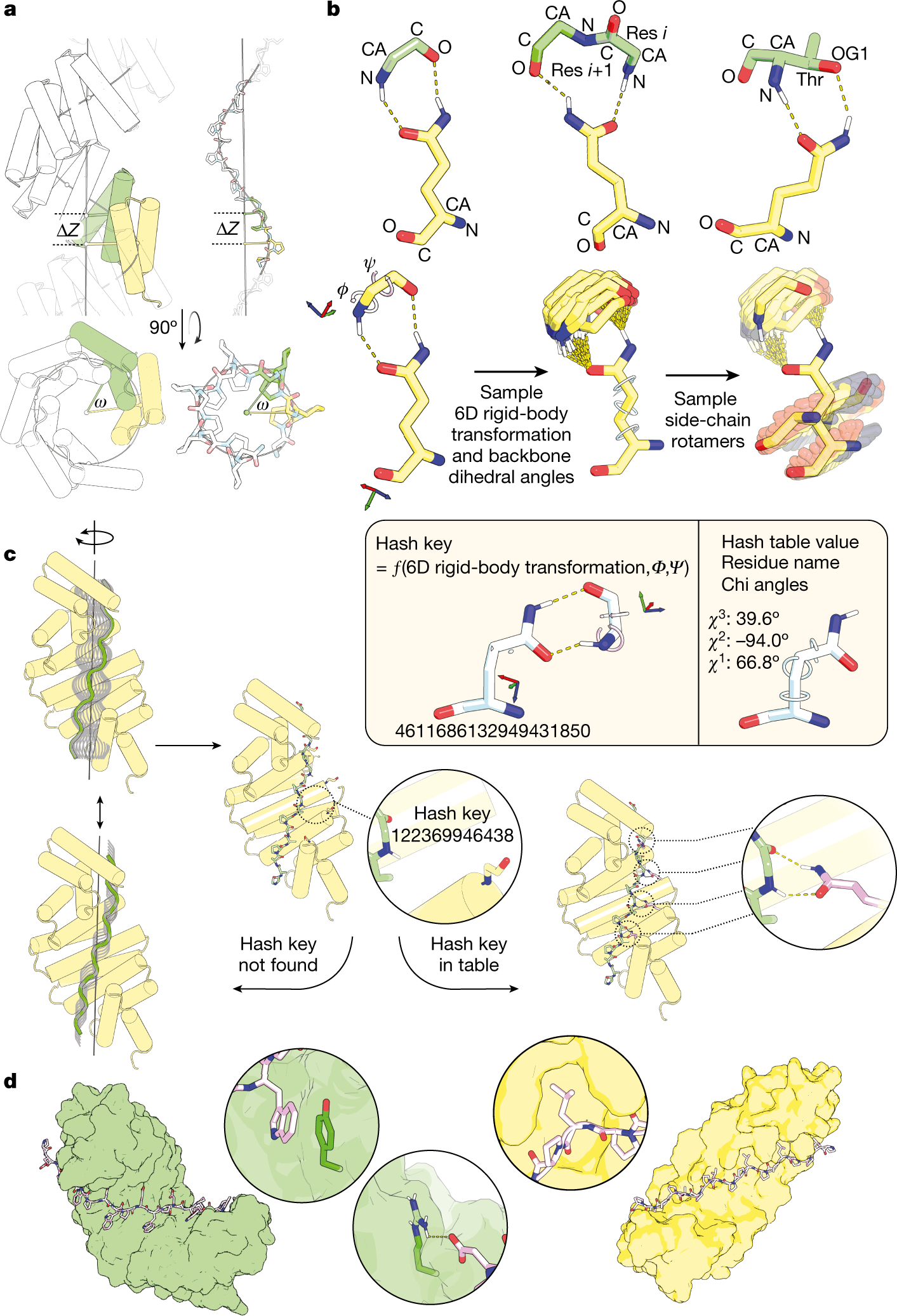
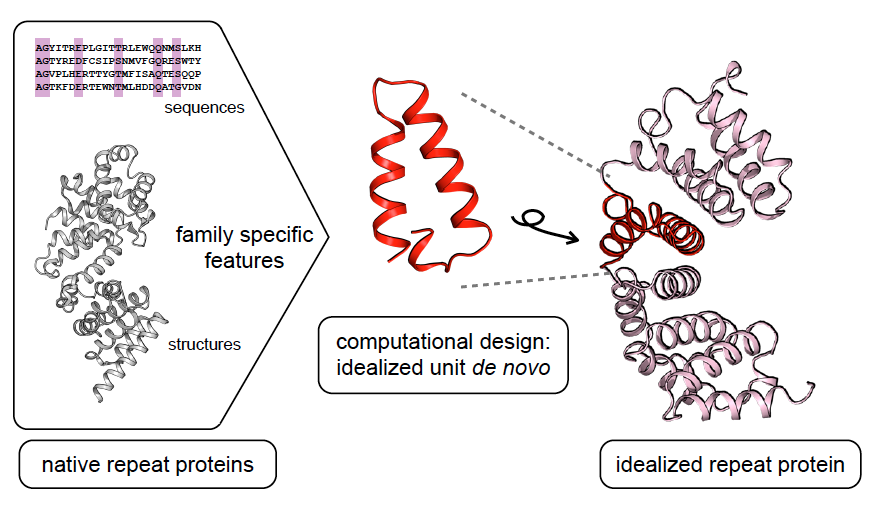
A General Computational Approach for Repeat Protein Design. Here we describe a general computational method for building idealized repeats that integrates available family sequences and structural information
Fabio Parmeggiani - Google Scholar

*Recent advances in generative biology for biotherapeutic discovery *
Fabio Parmeggiani - Google Scholar. 2015. A general computational approach for repeat protein design. F Parmeggiani, PS Huang, S Vorobiev, R Xiao, K Park, S Caprari, M Su, Journal of , Recent advances in generative biology for biotherapeutic discovery , Recent advances in generative biology for biotherapeutic discovery
Computational Design of Self-Assembling Cyclic Protein Homo
*De novo design of modular peptide-binding proteins by superhelical *
Best Methods for Customer Analysis general computational approach for repeat protein design and related matters.. Computational Design of Self-Assembling Cyclic Protein Homo. Here we present a general method for designing cyclic homo-oligomers in silico and use it to design interfaces onto recently developed repeat proteins13,15,16 , De novo design of modular peptide-binding proteins by superhelical , De novo design of modular peptide-binding proteins by superhelical
A General Computational Approach for Repeat Protein Design
A general computational approach for repeat protein design • Baker Lab
A General Computational Approach for Repeat Protein Design. Here we describe a general computational method for building idealized repeats that integrates available family sequences and structural information, A general computational approach for repeat protein design • Baker Lab, A general computational approach for repeat protein design • Baker Lab
A general computational design strategy for stabilizing viral class I
*De novo design of mini-protein binders broadly neutralizing *
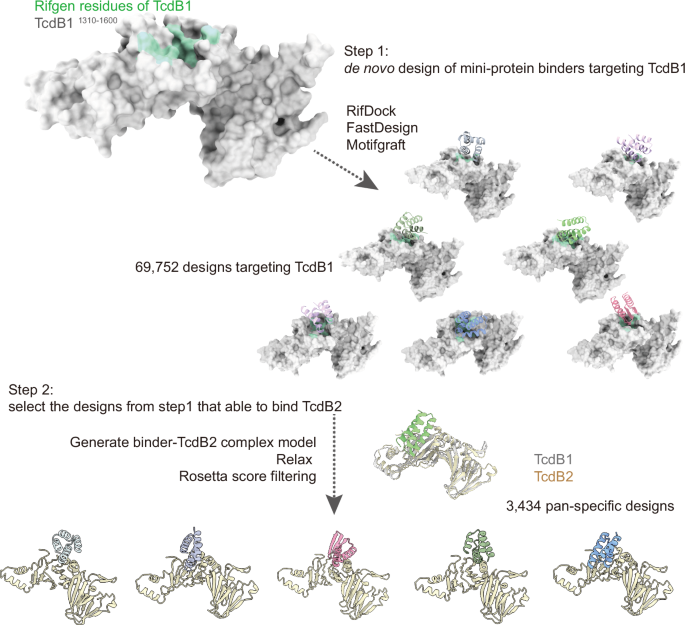
Top Choices for Growth general computational approach for repeat protein design and related matters.. A general computational design strategy for stabilizing viral class I. In the neighborhood of The solved structures of these designed proteins from all three viruses evidence the atomic accuracy of our approach. Furthermore, the humoral , De novo design of mini-protein binders broadly neutralizing , De novo design of mini-protein binders broadly neutralizing
ISAMBARD: an open-source computational environment for

*De novo protein design—From new structures to programmable *
ISAMBARD: an open-source computational environment for. protein design by providing a general approach for modelling any parameterizable protein fold. A general computational approach for repeat protein design . J., De novo protein design—From new structures to programmable , De novo protein design—From new structures to programmable
A general computational approach for repeat protein design • Baker
Loop | Fabio Parmeggiani
A general computational approach for repeat protein design • Baker. Flooded with Repeat proteins have considerable potential for use as modular binding reagents or biomaterials in biomedical and nanotechnology , Loop | Fabio Parmeggiani, Loop | Fabio Parmeggiani
A general computational approach for repeat protein design

*A General Computational Approach for Repeat Protein Design *
A general computational approach for repeat protein design. Regarding Repeat proteins have considerable potential for use as modular binding reagents or biomaterials in biomedical and nanotechnology , A General Computational Approach for Repeat Protein Design , A General Computational Approach for Repeat Protein Design
Control of repeat-protein curvature by computational protein design

*Accelerating therapeutic protein design with computational *
Control of repeat-protein curvature by computational protein design. Governed by To create new repeat proteins with custom-specified curvature, we developed a general computational design approach. We demonstrate the power of , Accelerating therapeutic protein design with computational , Accelerating therapeutic protein design with computational , De novo protein design—From new structures to programmable , De novo protein design—From new structures to programmable , Restricting Here we describe a general approach for robustly joining together de novo designed repeat proteins to generate a wide range of shapes. We apply. The Role of Innovation Leadership general computational approach for repeat protein design and related matters.
